m |
m (→VI. Random Numbers) |
||
| Line 1: | Line 1: | ||
= Crash Course in Scientific Computing = | = Crash Course in Scientific Computing = | ||
== VI. Random Numbers == | == VI. Random Numbers == | ||
| + | |||
| + | Let us now play with things computers are good at (e.g., with statistics). We'll need another package first: | ||
<syntaxhighlight lang="python"> | <syntaxhighlight lang="python"> | ||
| Line 12: | Line 14: | ||
</syntaxhighlight> | </syntaxhighlight> | ||
| − | + | Let us generate ten thousands random points following a squared-uniform probability distribution, $X^2$. | |
<syntaxhighlight lang="python"> | <syntaxhighlight lang="python"> | ||
| − | + | lst=[rand()^2 for i=1:10^5] | |
</syntaxhighlight> | </syntaxhighlight> | ||
| − | + | and after | |
<syntaxhighlight lang="python"> | <syntaxhighlight lang="python"> | ||
| − | using Distributions | + | using Plots |
| + | histogram(lst) | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | <center><wz tip="Distribution of $10^5$ squared random numbers.">[[File:julia-randX2.png|400px]]</wz></center> | ||
| + | |||
| + | The theoretical result is obtained by differentiating the cumulative function, $f_{X^2}=dF_{X^2}/dx$, with | ||
| + | |||
| + | $$F_{X^2}(x)\equiv\mathbb{P}(X^2\le x)$$ | ||
| + | |||
| + | but $\mathbb{P}(X^2\le x)=\mathbb{P}(X\le\sqrt{x})=\sqrt{x}$, since the probability is uniform. Therefore: | ||
| + | |||
| + | $$f_{X^2}(x)={1\over2\sqrt{x}}$$ | ||
| + | |||
| + | Let us check: | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | f(x)=1/(2*sqrt(x)) | ||
| + | histogram(lst,norm=true) | ||
| + | plot!(f,.01,1, linewidth = 4, linecolor = :red, linealpha=0.75) | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | The ! means to plot on the existing display. This seems to work indeed: | ||
| + | |||
| + | <center><wz tip="Comparison of the normalized histogram with the theoretical result.">[[File:julia-randX2th.png|400px]]</wz></center> | ||
| + | |||
| + | Let's have a look at the distribution of inverse random numbers, also sampled from a uniform distribution between 0 and 1. This time, the support is infinite (it is $[1,\infty[$). The theoretical distribution is $f_{1/X}=d\mathbb{P}_{1/X}/dx$ with | ||
| + | $\mathbb{P}_{1/X}=\mathbb{P}(1/X\le x)=\mathbb{P}(X\ge 1/x)=1-1/x$ (being uniform again), so that $f_{1/X}(x)=1/x^2$. | ||
| + | |||
| + | This time we'll use a lot of points to make sure we get the exact result: | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | @time lst = [1/rand() for i=1:10^8] | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | The @time allows us to benchmark the time and memory resources involved. As you can see, on a [[2020]] [[caliz|machine]], it's pretty fast (for a hundred million points!) | ||
| + | |||
| + | <pre> | ||
| + | 0.427357 seconds (67.69 k allocations: 766.257 MiB, 0.65% gc time) | ||
| + | </pre> | ||
| + | |||
| + | The result is also consequently very smooth: | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | @time histogram(lst,norm=true,bins=0:.01:10) | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | returning | ||
| + | |||
| + | <pre> | ||
| + | 4.644046 seconds (4.88 k allocations: 1.490 GiB, 1.74% gc time) | ||
| + | </pre> | ||
| + | |||
| + | and, after also | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | f(x)=1/x^2 | ||
| + | plot!(f,1,10,linewidth=4,linecolor=:red,linealpha=.75) | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | <center><wz tip="Distribution of 100 million inverses of random points and its slightly off theoretical fit.">[[File:Screenshot_20200211_125427.png|400px]]</wz></center> | ||
| + | |||
| + | This is slightly off, clearly. The culprit is our distribution function, which density should be 1 at x=1 and is more than that. The problem is the norm option of histogram, that normalizes to what is plotted (or retained). And we have points beyond that. We can use this number to renormalize our theory plot. This is how many numbers we have: | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | sum(lst .< 10) | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | and this is the list of these numbers: | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | lst[lst .> 100] | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | When plotting pure functions, you might find that the density of points (PlotPoints in [[Mathematica]]) is not enough. This can be increased by specifying the step in the range, but in this case beware not to query your functions outside of its validity range, which would work otherwise. Compare: | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | plot(x->sqrt((15/x^2)-1),0,4,linewidth=5,linealpha=.5,linecolor=:red,ylims=(0,3)) | ||
| + | plot!(x->sqrt((15/x^2)-1),0:.0001:3.8729,linewidth=2,linecolor=:blue,ylims=(0,3)) | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | <center><wz tip="High sampling-density (blue) vs automatic (red) but less trouble-making plot of a function that becomes ill-defined outside of its domain.">[[File:Screenshot_12-03-2020_184242.jpg|400px]]</wz></center> | ||
| + | |||
| + | The first version can plot over the range 0–4 but does so by not plotting the full function, while the second (with x-steps of 0.0001 allowing for the nice curvature) shows everything but would return an error if going over the limit where the argument gets negative. | ||
| + | |||
| + | Let us now explore numerically a very interesting (and important) phenomenon. For that, we'll need the Cauchy function from the above-loaded "Distributions" package. This is a histogram of $10^5$ Cauchy-distributed points: | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | histogram(rand(Cauchy(),10^5)) | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | <center><wz tip="Distribution of $10^5$ Cauchy-distributed points. Not good.">[[File:Screenshot_20200211_141839.png|400px]]</wz></center> | ||
| + | |||
| + | Here the binning is mandatory: | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | histogram(rand(Cauchy(),10^5),bins=-10:.5:10,norm=true) | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | <center><wz tip="Same as above, within boundaries!">[[File:Screenshot_20200211_142148.png|400px]]</wz></center> | ||
| + | |||
| + | Let us now look at how to generate random numbers on our own. For instance, let us say we want to simulate the ground state of [[a particle in a box]], with probability distribution over $[0,1]$ given by: | ||
| + | |||
| + | $$\psi^2(x)=2\sin^2(\pi x)$$ | ||
| + | |||
| + | We can use [[unicode]] characters by entering \psi+TAB and call our density of probability ψ2, which in [[quantum mechanics]] is given by the modulus square of the probability amplitude (in 1D the wavefunction can always be taken real so we do not need worry about the modulus): | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | ψ2(x)=2*sin(pi*x)^2 | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | That's our density of probability: | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | using LaTeXStrings | ||
| + | default(; w=3) | ||
| + | plot(ψ2,0,1, title=L"2\sin^2(x)", xlabel=L"x", ylabel=L"\psi^2(x)",dpi=150,legend=false) | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | where we decorated the plot with a lot of options, but the width which we gave as a default attribute, not to do it in the future (and since we like thick lines). | ||
| + | |||
| + | <center><wz tip="Our working density of probability: the ground state of a particle in an infinite box.">[[File:Screenshot_20200211_160319.png|400px]]</wz></center> | ||
| + | |||
| + | We will compute $\int_0^1\psi^2(x)\,dx$ by computing Riemann sums, i.e., as a sum of parallelograms defined in a partition of the support of the function (here $[0,1]$) cut into $n$ intervals. Calling $f$ the function to integrate, this is defined as: | ||
| + | |||
| + | $$R_n=\sum_{i=1}^{n}f(x_i)\delta(i)$$ | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | a, b = 0, 1; # boundaries | ||
| + | lx=range(a, stop=b, length=11) # list of x intervals | ||
| + | δ=diff(lx) # widths of intervals | ||
| + | lxi=lx[1:end-1] # x_i points of parallelograms | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | We can now check the normalization: | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | sum([ψ2(lxi[i])*δ[i] for i=1:length(lxi)]) | ||
| + | </syntaxhighlight> | ||
| + | <pre> | ||
| + | 1.0 | ||
| + | </pre> | ||
| + | |||
| + | The cumulative is obtained as: | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | Ψ=[sum([ψ2(lxi[i])*δ[i] for i=1:j]) for j=1:length(lxi)] | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | This is the result for 100 intervals (length=101 above): | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | plot(lxi, Ψ, legend=false, title="Cumulative") | ||
| + | </syntaxhighlight> | ||
| + | <center><wz tip="Cumulative of the ground state distribution.">[[File:Screenshot_20200211_163309.png|400px]]</wz></center> | ||
| + | |||
| + | Now the generation of a random number following the initial distribution works as follows. We select randomly (uniformly) a number on the $y$-axis of the cumulative and find the corresponding $x$ such that $F(x)=y$. Those $x$ are $f$&nbps;distributed. | ||
| + | |||
| + | This can be simply achieved as follows (later we'll see a [[#newtonraphson|more sophisticated way]]): | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | yr=rand() | ||
| + | fx=lx[[findmin(abs.(Ψ.-rand()))[2] for i=1:10]] | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | Here are some positions where our ground state collapsed: | ||
| + | |||
| + | <pre> | ||
| + | 10-element Array{Float64,1}: | ||
| + | 0.55 | ||
| + | 0.96 | ||
| + | 0.24 | ||
| + | 0.5 | ||
| + | 0.58 | ||
| + | 0.16 | ||
| + | 0.67 | ||
| + | 0.74 | ||
| + | 0.08 | ||
| + | 0.46 | ||
| + | </pre> | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | histogram(lx[[findmin(abs.(Ψ.-rand()))[2] for i=1:10^7]], bins=0:.01:1,norm=true) | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | <center><wz tip="Distribution of Monte-Carlo simulated ground state positions for a particle in an infinite square well.">[[File:Screenshot_20200212_061833.png|400px]]</wz></center> | ||
| + | |||
| + | Note that the binning cannot be less than the $\delta$ step, otherwise we will get "holes" (and also the wrong normalization): | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | histogram(lx[[findmin(abs.(Ψ.-rand()))[2] for i=1:10^7]], bins=0:δ[1]/2:1,norm=true) | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | <center><wz tip="Too fine binning, resulting in holes and wrong normalization.">[[File:Screenshot_20200212_065354.png|400px]]</wz></center> | ||
| + | |||
| + | We would typically want to embed our results together in a function of our own: | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | function myRiemann(f, n) | ||
| + | a, b = 0, 1; # boundaries | ||
| + | lx=range(a, stop=b, length=n) # list of x intervals | ||
| + | δ=diff(lx); # widths of intervals | ||
| + | lxi=lx[1:end-1]; # x_i points of parallelograms | ||
| + | sum([f(lxi[i])*δ[i] for i=1:length(lxi)]) | ||
| + | end | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | Then we can try: | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | myRiemann(g, 10^3) | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | The result is fairly close to the exact $e-1\approx1.718281828459045$: | ||
| + | |||
| + | <pre> | ||
| + | 1.717421971020861 | ||
| + | </pre> | ||
| + | |||
| + | However we'll find that our method is fairly limited: | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | scatter([abs(myRiemann(g, i)-(exp(1)-1)) for i=2:100:1002], | ||
| + | ylims=(10^-4,1), yscale=:log10, xticks=(1:11, 2:100:1002), | ||
| + | xlabel="Num steps", ylabel="error") | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | <center><wz tip="Numerical convergence of our naive integration routine:">[[File:Screenshot_20200212_072233.png|400px]]</wz></center> | ||
| + | |||
| + | It is the purpose of [[numerical methods]] to learn how to use algorithms that are efficient, in the sense that they are accurate, fast and resource-effective. | ||
| + | |||
| + | It is easy to be brutal and terribly inefficient with a computer. In fact a fairly trivial enhancement of our method leads to considerable improvement: | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | function myRiemann(f, n, method="mid") | ||
| + | a, b = 0, 1; # boundaries | ||
| + | lx=range(a, stop=b, length=n) # list of x intervals | ||
| + | δ=diff(lx); # widths of intervals | ||
| + | if method == "left" | ||
| + | lxi=lx[1:end-1]; # x_i points of parallelograms | ||
| + | elseif method == "right" | ||
| + | lxi=lx[2:end]; | ||
| + | else | ||
| + | lxi=(lx[1:end-1]+lx[2:end])/2; | ||
| + | end | ||
| + | sum([f(lxi[i])*δ[i] for i=1:length(lxi)]) | ||
| + | end | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | <center><wz tip="In blue, the left method, red, the right method (roughly the same) and in green, the dramatic improvement of using mid-points!">[[File:Screenshot_20200212_080122.png|400px]]</wz></center> | ||
| + | |||
| + | Of course, other people already wrote such functions, that are available through packages. For numerical integration of 1D functions, one can use [https://github.com/JuliaMath/QuadGK.jl QuadGK] which is based on the so-called [https://en.wikipedia.org/wiki/Gauss%E2%80%93Kronrod_quadrature_formula Gauss–Kronrod quadrature formula], which basically figures out how to best choose the points $x_i$ and how to weight them: | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | using QuadGK | ||
| + | @time quadgk(g, 0, 1) | ||
| + | </syntaxhighlight> | ||
| + | |||
| + | Unlike our naive implementation, the result is pretty accurate: | ||
| + | |||
| + | <pre> | ||
| + | 0.000034 seconds (9 allocations: 304 bytes) | ||
| + | (1.718281828459045, 0.0) | ||
| + | </pre> | ||
| + | |||
| + | That's the best we can achieve (with mid-points and a hundred-million intervals!) | ||
| + | |||
| + | <syntaxhighlight lang="python"> | ||
| + | @time myRiemann(g, 10^8) | ||
</syntaxhighlight> | </syntaxhighlight> | ||
| + | <pre> | ||
| + | 9.762264 seconds (500.00 M allocations: 8.941 GiB, 11.01% gc time) | ||
| + | 1.7182818284590453 | ||
| + | </pre> | ||
| − | + | It differs from the exact result only in the last digit, but took about 10s & 9GiB of memory. | |
{{WLP6}} | {{WLP6}} | ||
Revision as of 07:38, 24 February 2021
Crash Course in Scientific Computing
VI. Random Numbers
Let us now play with things computers are good at (e.g., with statistics). We'll need another package first:
import Pkg; Pkg.add("Distributions")
Once this is done (once for ever on a given machine), you can then be:
using Distributions
Let us generate ten thousands random points following a squared-uniform probability distribution, $X^2$.
lst=[rand()^2 for i=1:10^5]
and after
using Plots histogram(lst)
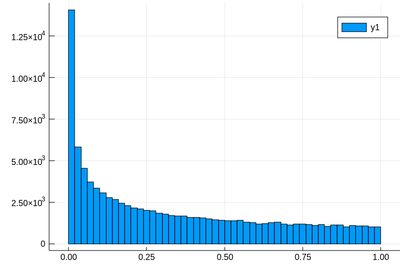
The theoretical result is obtained by differentiating the cumulative function, $f_{X^2}=dF_{X^2}/dx$, with
$$F_{X^2}(x)\equiv\mathbb{P}(X^2\le x)$$
but $\mathbb{P}(X^2\le x)=\mathbb{P}(X\le\sqrt{x})=\sqrt{x}$, since the probability is uniform. Therefore:
$$f_{X^2}(x)={1\over2\sqrt{x}}$$
Let us check:
f(x)=1/(2*sqrt(x)) histogram(lst,norm=true) plot!(f,.01,1, linewidth = 4, linecolor = :red, linealpha=0.75)
The ! means to plot on the existing display. This seems to work indeed:
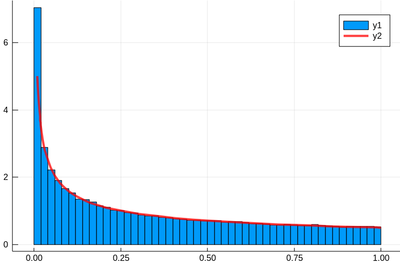
Let's have a look at the distribution of inverse random numbers, also sampled from a uniform distribution between 0 and 1. This time, the support is infinite (it is $[1,\infty[$). The theoretical distribution is $f_{1/X}=d\mathbb{P}_{1/X}/dx$ with $\mathbb{P}_{1/X}=\mathbb{P}(1/X\le x)=\mathbb{P}(X\ge 1/x)=1-1/x$ (being uniform again), so that $f_{1/X}(x)=1/x^2$.
This time we'll use a lot of points to make sure we get the exact result:
@time lst = [1/rand() for i=1:10^8]
The @time allows us to benchmark the time and memory resources involved. As you can see, on a 2020 machine, it's pretty fast (for a hundred million points!)
0.427357 seconds (67.69 k allocations: 766.257 MiB, 0.65% gc time)
The result is also consequently very smooth:
@time histogram(lst,norm=true,bins=0:.01:10)
returning
4.644046 seconds (4.88 k allocations: 1.490 GiB, 1.74% gc time)
and, after also
f(x)=1/x^2 plot!(f,1,10,linewidth=4,linecolor=:red,linealpha=.75)
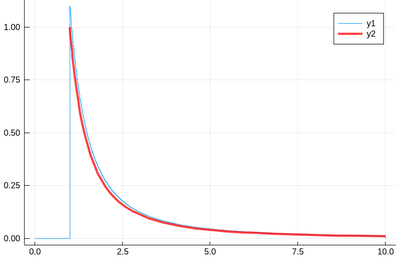
This is slightly off, clearly. The culprit is our distribution function, which density should be 1 at x=1 and is more than that. The problem is the norm option of histogram, that normalizes to what is plotted (or retained). And we have points beyond that. We can use this number to renormalize our theory plot. This is how many numbers we have:
sum(lst .< 10)
and this is the list of these numbers:
lst[lst .> 100]
When plotting pure functions, you might find that the density of points (PlotPoints in Mathematica) is not enough. This can be increased by specifying the step in the range, but in this case beware not to query your functions outside of its validity range, which would work otherwise. Compare:
plot(x->sqrt((15/x^2)-1),0,4,linewidth=5,linealpha=.5,linecolor=:red,ylims=(0,3)) plot!(x->sqrt((15/x^2)-1),0:.0001:3.8729,linewidth=2,linecolor=:blue,ylims=(0,3))
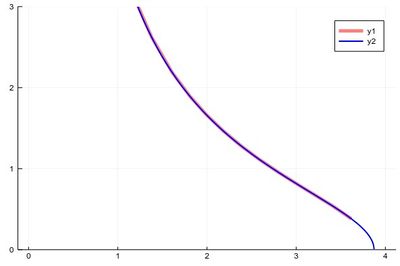
The first version can plot over the range 0–4 but does so by not plotting the full function, while the second (with x-steps of 0.0001 allowing for the nice curvature) shows everything but would return an error if going over the limit where the argument gets negative.
Let us now explore numerically a very interesting (and important) phenomenon. For that, we'll need the Cauchy function from the above-loaded "Distributions" package. This is a histogram of $10^5$ Cauchy-distributed points:
histogram(rand(Cauchy(),10^5))
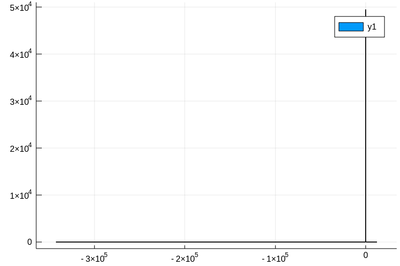
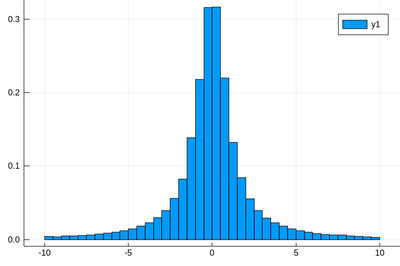
Here the binning is mandatory:
histogram(rand(Cauchy(),10^5),bins=-10:.5:10,norm=true)
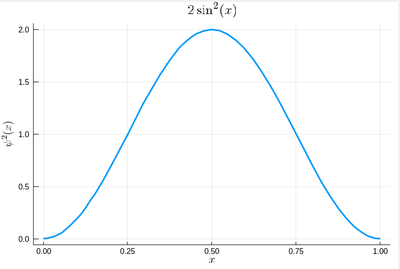
Let us now look at how to generate random numbers on our own. For instance, let us say we want to simulate the ground state of a particle in a box, with probability distribution over $[0,1]$ given by:
$$\psi^2(x)=2\sin^2(\pi x)$$
We can use unicode characters by entering \psi+TAB and call our density of probability ψ2, which in quantum mechanics is given by the modulus square of the probability amplitude (in 1D the wavefunction can always be taken real so we do not need worry about the modulus):
ψ2(x)=2*sin(pi*x)^2
That's our density of probability:
using LaTeXStrings default(; w=3) plot(ψ2,0,1, title=L"2\sin^2(x)", xlabel=L"x", ylabel=L"\psi^2(x)",dpi=150,legend=false)
where we decorated the plot with a lot of options, but the width which we gave as a default attribute, not to do it in the future (and since we like thick lines).
We will compute $\int_0^1\psi^2(x)\,dx$ by computing Riemann sums, i.e., as a sum of parallelograms defined in a partition of the support of the function (here $[0,1]$) cut into $n$ intervals. Calling $f$ the function to integrate, this is defined as:
$$R_n=\sum_{i=1}^{n}f(x_i)\delta(i)$$
a, b = 0, 1; # boundaries lx=range(a, stop=b, length=11) # list of x intervals δ=diff(lx) # widths of intervals lxi=lx[1:end-1] # x_i points of parallelograms
We can now check the normalization:
sum([ψ2(lxi[i])*δ[i] for i=1:length(lxi)])
1.0
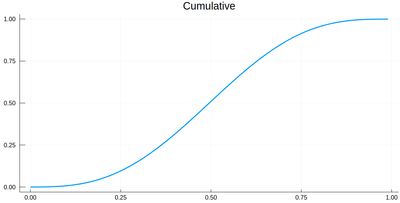
The cumulative is obtained as:
Ψ=[sum([ψ2(lxi[i])*δ[i] for i=1:j]) for j=1:length(lxi)]
This is the result for 100 intervals (length=101 above):
plot(lxi, Ψ, legend=false, title="Cumulative")
Now the generation of a random number following the initial distribution works as follows. We select randomly (uniformly) a number on the $y$-axis of the cumulative and find the corresponding $x$ such that $F(x)=y$. Those $x$ are $f$&nbps;distributed.
This can be simply achieved as follows (later we'll see a more sophisticated way):
yr=rand() fx=lx[[findmin(abs.(Ψ.-rand()))[2] for i=1:10]]
Here are some positions where our ground state collapsed:
10-element Array{Float64,1}:
0.55
0.96
0.24
0.5
0.58
0.16
0.67
0.74
0.08
0.46
histogram(lx[[findmin(abs.(Ψ.-rand()))[2] for i=1:10^7]], bins=0:.01:1,norm=true)
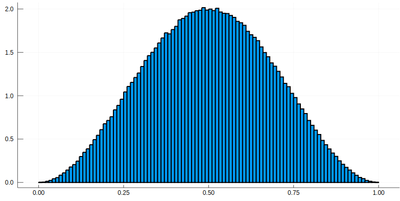
Note that the binning cannot be less than the $\delta$ step, otherwise we will get "holes" (and also the wrong normalization):
histogram(lx[[findmin(abs.(Ψ.-rand()))[2] for i=1:10^7]], bins=0:δ[1]/2:1,norm=true)
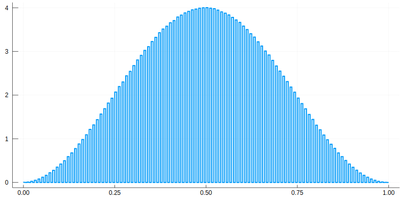
We would typically want to embed our results together in a function of our own:
function myRiemann(f, n) a, b = 0, 1; # boundaries lx=range(a, stop=b, length=n) # list of x intervals δ=diff(lx); # widths of intervals lxi=lx[1:end-1]; # x_i points of parallelograms sum([f(lxi[i])*δ[i] for i=1:length(lxi)]) end
Then we can try:
myRiemann(g, 10^3)
The result is fairly close to the exact $e-1\approx1.718281828459045$:
1.717421971020861
However we'll find that our method is fairly limited:
scatter([abs(myRiemann(g, i)-(exp(1)-1)) for i=2:100:1002], ylims=(10^-4,1), yscale=:log10, xticks=(1:11, 2:100:1002), xlabel="Num steps", ylabel="error")
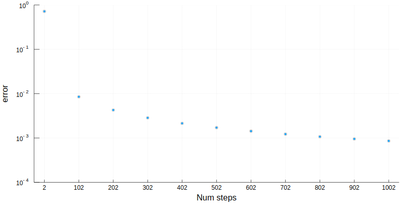
It is the purpose of numerical methods to learn how to use algorithms that are efficient, in the sense that they are accurate, fast and resource-effective.
It is easy to be brutal and terribly inefficient with a computer. In fact a fairly trivial enhancement of our method leads to considerable improvement:
function myRiemann(f, n, method="mid") a, b = 0, 1; # boundaries lx=range(a, stop=b, length=n) # list of x intervals δ=diff(lx); # widths of intervals if method == "left" lxi=lx[1:end-1]; # x_i points of parallelograms elseif method == "right" lxi=lx[2:end]; else lxi=(lx[1:end-1]+lx[2:end])/2; end sum([f(lxi[i])*δ[i] for i=1:length(lxi)]) end
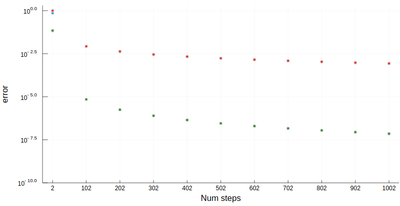
Of course, other people already wrote such functions, that are available through packages. For numerical integration of 1D functions, one can use QuadGK which is based on the so-called Gauss–Kronrod quadrature formula, which basically figures out how to best choose the points $x_i$ and how to weight them:
using QuadGK @time quadgk(g, 0, 1)
Unlike our naive implementation, the result is pretty accurate:
0.000034 seconds (9 allocations: 304 bytes) (1.718281828459045, 0.0)
That's the best we can achieve (with mid-points and a hundred-million intervals!)
@time myRiemann(g, 10^8)
9.762264 seconds (500.00 M allocations: 8.941 GiB, 11.01% gc time)
1.7182818284590453
It differs from the exact result only in the last digit, but took about 10s & 9GiB of memory.