m (→XIII. Integrals) |
m (→XIII. Integrals) |
||
| Line 155: | Line 155: | ||
A great choice of interpolating polynomials is the so-called Lagrange-interpolating polynomials, which provide the unique polynomial of order $n$ to pass by the $n$ points $(x_i, y_i)$ (with $1\le i\le n$). Note that the order of the polynomial is the same as the number of points one wishes to pass through. These polynomials are obtained as a linear superposition of the Lagrange-basis polynomials: | A great choice of interpolating polynomials is the so-called Lagrange-interpolating polynomials, which provide the unique polynomial of order $n$ to pass by the $n$ points $(x_i, y_i)$ (with $1\le i\le n$). Note that the order of the polynomial is the same as the number of points one wishes to pass through. These polynomials are obtained as a linear superposition of the Lagrange-basis polynomials: | ||
| − | $$\ell _{j}(x)\equiv\prod _{\begin{smallmatrix}1\leq m\leq | + | $$\ell _{j}(x)\equiv\prod _{\begin{smallmatrix}1\leq m\leq n\\m\neq j\end{smallmatrix}}{\frac {x-x_{m}}{x_{j}-x_{m}}}={\frac {(x-x_{1})}{(x_{j}-x_{1})}}\cdots {\frac {(x-x_{j-1})}{(x_{j}-x_{j-1})}}{\frac {(x-x_{j+1})}{(x_{j}-x_{j+1})}}\cdots {\frac {(x-x_{n})}{(x_{j}-x_{n})}}$$ |
They are such that $l_j(x_k)=0$ for all $1\le k\le n$ such that $k\neq j$ where $l_j(x_k)=1$. | They are such that $l_j(x_k)=0$ for all $1\le k\le n$ such that $k\neq j$ where $l_j(x_k)=1$. | ||
Revision as of 09:40, 12 March 2021
Crash Course in Scientific Computing
XIII. Integrals
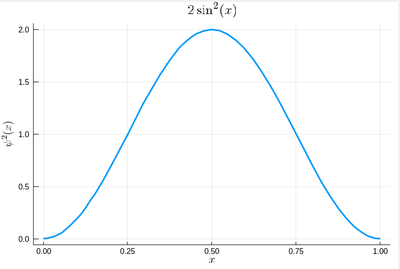
We will now start to apply algorithms, or precise recipes, to numerical problems. We will start with a simple one, that has a clear physical interpretation: integration of 1D functions of one variable, i.e., the computation of areas. We will also look at a precise application of such computations (which are of interest on their own, of course). Namely, we will look at how to generate random numbers on our own, that is, following any given probability distribution (a positive, normalized function). For instance, let us say we want to simulate the ground state of a particle in a box, with probability distribution over $[0,1]$ given by:
$$\psi^2(x)=2\sin^2(\pi x)$$
We remind that one can use unicode characters (by entering \psi+TAB) to call our density of probability ψ2, which in quantum mechanics is given by the modulus square of the probability amplitude (in 1D the wavefunction can always be taken real so we do not need worry about the modulus):
ψ2(x)=2*sin(pi*x)^2
That's our density of probability:
using LaTeXStrings default(; w=3) plot(ψ2,0,1, title=L"2\sin^2(x)", xlabel=L"x", ylabel=L"\psi^2(x)",dpi=150,legend=false)
where we decorated the plot with a lot of options, but the width which we gave as a default attribute, not to do it in the future (and since we like thick lines).
To draw random numbers that follow this probability distribution, we will need to compute quantities such as $F(y)\equiv\int_0^y\psi^2(x)\,dx$, or the so-called cumulative. Of course we should find that $F(1)=1$ (normalization condition), so we can check that as well. The simplest way to compute such integrals numerically is to bring to the computer the mathematical definition we have given of the integral, namely, in terms of Riemann sums, i.e., as a sum of parallelograms defined on a partition of the support of the function (here $[0,1]$) cut into $n$ intervals. Calling $f$ the function to integrate, this is defined as:
$$R_n=\sum_{i=1}^{n}f(x_i)\delta(i)$$
a, b = 0, 1; # boundaries lx=range(a, stop=b, length=11) # list of x intervals δ=diff(lx) # widths of intervals lxi=lx[1:end-1] # x_i points of parallelograms
We can now compute our integral over the entire interval to check the normalization:
sum([ψ2(lxi[i])*δ[i] for i=1:length(lxi)])
1.0
The cumulative is itself obtained as:
Ψ=[sum([ψ2(lxi[i])*δ[i] for i=1:j]) for j=1:length(lxi)]
This is the result for 100 intervals (length=101 above):
plot(lxi, Ψ, legend=false, title="Cumulative")
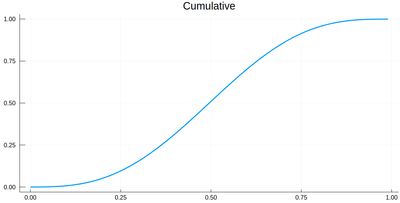
Now the generation of a random number following the initial distribution works as follows. We select randomly (uniformly) a number on the $y$-axis of the cumulative and find the corresponding $x$ such that $F(x)=y$. Those $x$ are $f$ distributed.
This can be simply achieved as follows (later we'll see a more sophisticated way but let's just use the most straightforward way to get to the point):
yr=rand() fx=lx[[findmin(abs.(Ψ.-rand()))[2] for i=1:10]]
Here are some positions where our ground state collapsed:
10-element Array{Float64,1}:
0.55
0.96
0.24
0.5
0.58
0.16
0.67
0.74
0.08
0.46
histogram(lx[[findmin(abs.(Ψ.-rand()))[2] for i=1:10^7]], bins=0:.01:1,norm=true)
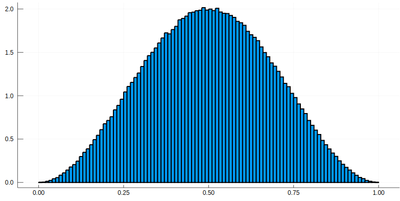
Note that the binning cannot be less than the $\delta$ step, otherwise we will get "holes" (and also the wrong normalization) (?!)Too fine binning, resulting in holes and wrong normalization.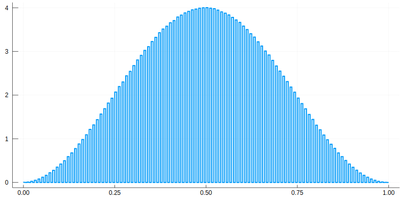 .
.
Putting our results together in a function of our own:
function myRiemann(f, n) a, b = 0, 1; # boundaries lx=range(a, stop=b, length=n) # list of x intervals δ=diff(lx); # widths of intervals lxi=lx[1:end-1]; # x_i points of parallelograms sum([f(lxi[i])*δ[i] for i=1:length(lxi)]) end
Then we can try:
myRiemann(g, 10^3)
The result is fairly close to the exact $e-1\approx1.718281828459045$:
1.717421971020861
However we'll find that our method is fairly limited:
scatter([abs(myRiemann(g, i)-(exp(1)-1)) for i=2:100:1002], ylims=(10^-4,1), yscale=:log10, xticks=(1:11, 2:100:1002), xlabel="Num steps", ylabel="error")
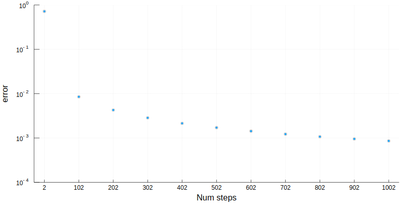
It is the purpose of numerical methods to learn how to use algorithms that are efficient, in the sense that they are accurate, fast and resource-effective.
It is easy to be brutal and terribly inefficient with a computer. In fact a fairly trivial enhancement of our method leads to considerable improvement. We have used Riemann integrals by taking the leftmost point. We could also take the rightmost point, or the central point, i.e., their average:
function myRiemann(f, n, method="mid") a, b = 0, 1; # boundaries lx=range(a, stop=b, length=n) # list of x intervals δ=diff(lx); # widths of intervals if method == "left" lxi=lx[1:end-1]; # x_i points of parallelograms elseif method == "right" lxi=lx[2:end]; else lxi=(lx[1:end-1]+lx[2:end])/2; end sum([f(lxi[i])*δ[i] for i=1:length(lxi)]) end
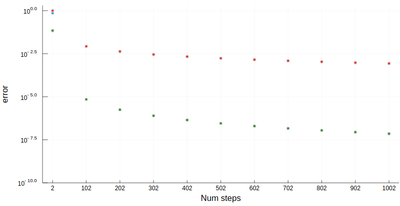
There is a huge gain in precision by evaluating the function elsewhere? Why? Clearly because we find a compensation of errors. Note that evaluating at the central point is equivalent to computing the area of a trapezoid, which also explain why the area is better estimated. For this reason, this method of integration is called the "trapezoidal rule".
Can it be further improved? Of course, following the same idea, one could approximate the function not by a straight line, but by a curve. Polynomials make easy curves and they are easy to integrate too, not even numerically, but symbolically. That brings forward the so-called "Newton-Cotes quadrature" that specify the weights $\omega_i$ and the locations $x_i$ where to sample the function to integrate:
$$\int _{a}^{b}f(x)\,dx\approx \sum _{i=0}^{n}w_{i}\,f(x_{i})$$
A great choice of interpolating polynomials is the so-called Lagrange-interpolating polynomials, which provide the unique polynomial of order $n$ to pass by the $n$ points $(x_i, y_i)$ (with $1\le i\le n$). Note that the order of the polynomial is the same as the number of points one wishes to pass through. These polynomials are obtained as a linear superposition of the Lagrange-basis polynomials:
$$\ell _{j}(x)\equiv\prod _{\begin{smallmatrix}1\leq m\leq n\\m\neq j\end{smallmatrix}}{\frac {x-x_{m}}{x_{j}-x_{m}}}={\frac {(x-x_{1})}{(x_{j}-x_{1})}}\cdots {\frac {(x-x_{j-1})}{(x_{j}-x_{j-1})}}{\frac {(x-x_{j+1})}{(x_{j}-x_{j+1})}}\cdots {\frac {(x-x_{n})}{(x_{j}-x_{n})}}$$
They are such that $l_j(x_k)=0$ for all $1\le k\le n$ such that $k\neq j$ where $l_j(x_k)=1$.
Of course, other people already wrote such functions, that are available through packages. For numerical integration of 1D functions, one can use QuadGK which is based on the so-called Gauss–Kronrod quadrature formula, which basically figures out how to best choose the points $x_i$ and how to weight them:
using QuadGK @time quadgk(g, 0, 1)
Unlike our naive implementation, the result is pretty accurate:
0.000034 seconds (9 allocations: 304 bytes) (1.718281828459045, 0.0)
That's the best we can achieve (with mid-points and a hundred-million intervals!)
@time myRiemann(g, 10^8)
9.762264 seconds (500.00 M allocations: 8.941 GiB, 11.01% gc time)
1.7182818284590453
It differs from the exact result only in the last digit, but took about 10s & 9GiB of memory.